Motivation
The current availability of public biomedical data and ontologies can be leveraged towards the development of explainable knowledge enabled systems in biomedical Artificial Intelligence, with the objective of developing personalized medicine solutions to aid clinicians and researchers.
We propose an approach to build a Knowledge Graph (KG) for personalized medicine to serve as a rich input for the AI system and incorporate its outcomes to support explanations, by connecting input and output.
Personalized Medicine
is a data-oriented medical model that relies on the specific characteristics of the patient and thus encompasses a very diverse and complex domain

Explainable Artificial Intelligence (XAI)
is able to handle the vast biomedical data pool available and has the goal of providing human-understandable explanations in order to increase trust in AI model predictions

Knowledge Graphs
are able to combine biomedical ontologies and data to provide a rich and complex domain context, functioning as input for biomedical XAI applications capable of producing explanations
Our approach
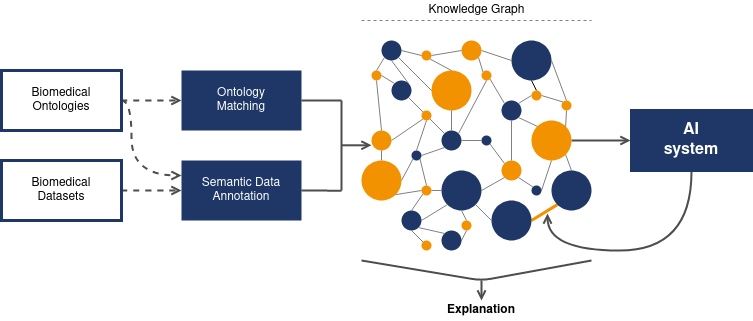
Resource curation
Selection and curation of relevant public biomedical datasets and ontologies
Challenges:
- Adequate coverage of the domain
- Semantic richness for explanations
Ontology Matching
Ontologies will be matched iteratively and alignments will be partially validated by experts
Challenges:
- Scalability
- Complex matching
Semantic Data Annotation
Development of parsers and annotation algorithms
Challenges:
- Diverse types of datasets
AI Integration
Ensure that the KG serves as both input to AI methods and also encodes the AI outcomes
Challenges:
- Diverse types of AI outcomes
Conclusions
- A biomedical KG obtained from the intersection of biomedical ontologies and data can be leveraged as input to AI methods and can encode its outcomes
- Such a KG can support a shared semantic space for data, scientific context, and predictions capable of supporting KG-based explanation methods
Funding
This work was supported by FCT through the LASIGE Research Unit (UIDB/00408/2020 and UIDP/00408/2020). It was also partially supported by the KATY project which has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 101017453.






