Motivation
In the biomedical field, Machine Learning and Artificial Intelligence models have shown impressive successes, but they often lack some degree of explainability, in the sense they fail to provide human-understandable logical decisions.
Discovering Disease Genes (DGs) is one particular task in the biomedical research field that requires some degree of explainability, particularly for prioritizing genes for further research.
Background
S2B is a network-based method that uses protein-protein interaction (PPI) networks to predict DGs associated with two similar diseases.
In this case, it’s important to understand why certain genes may be related with two similar diseases, and adding knowledge to the process may improve gene prioritization.
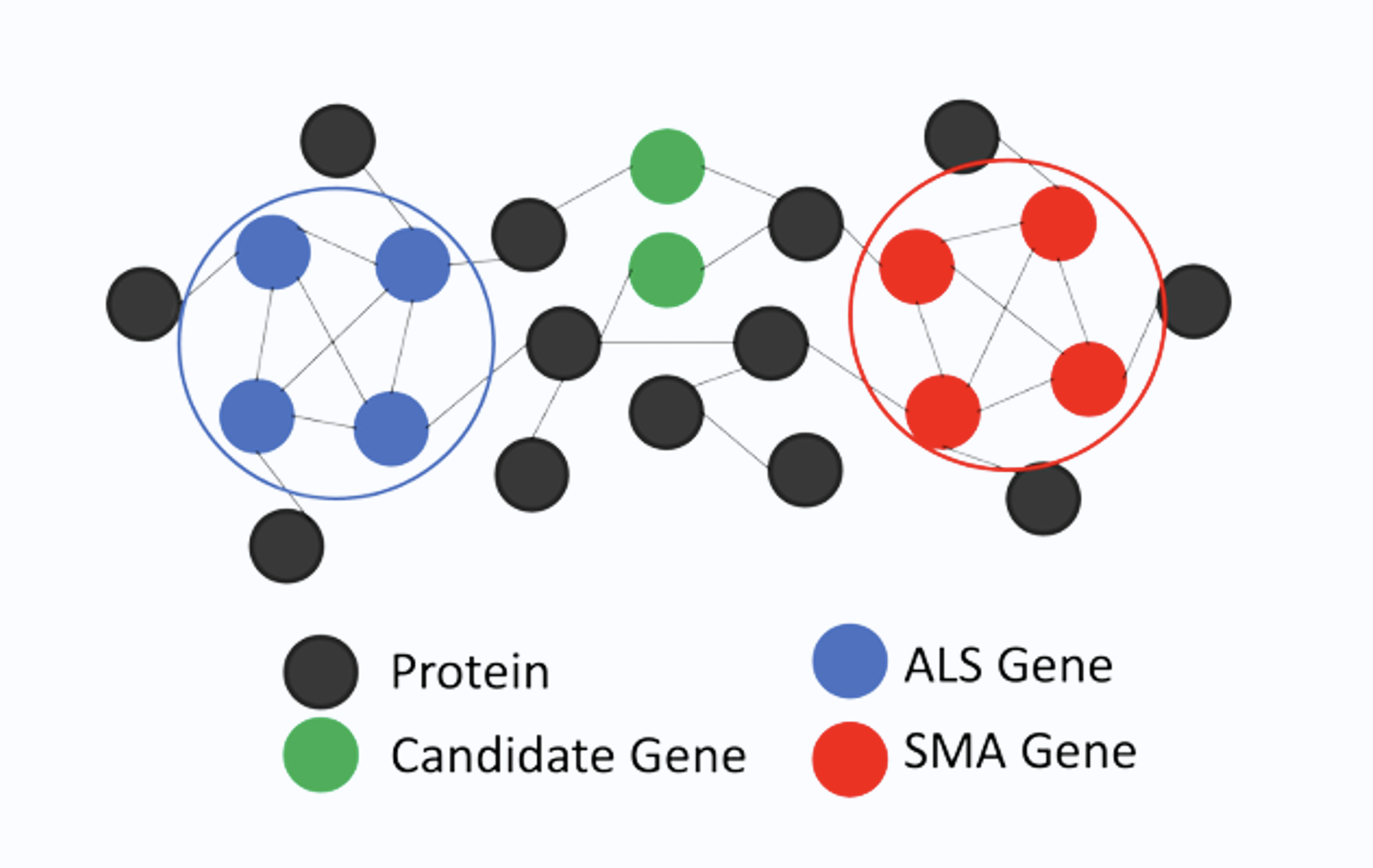
Methodology
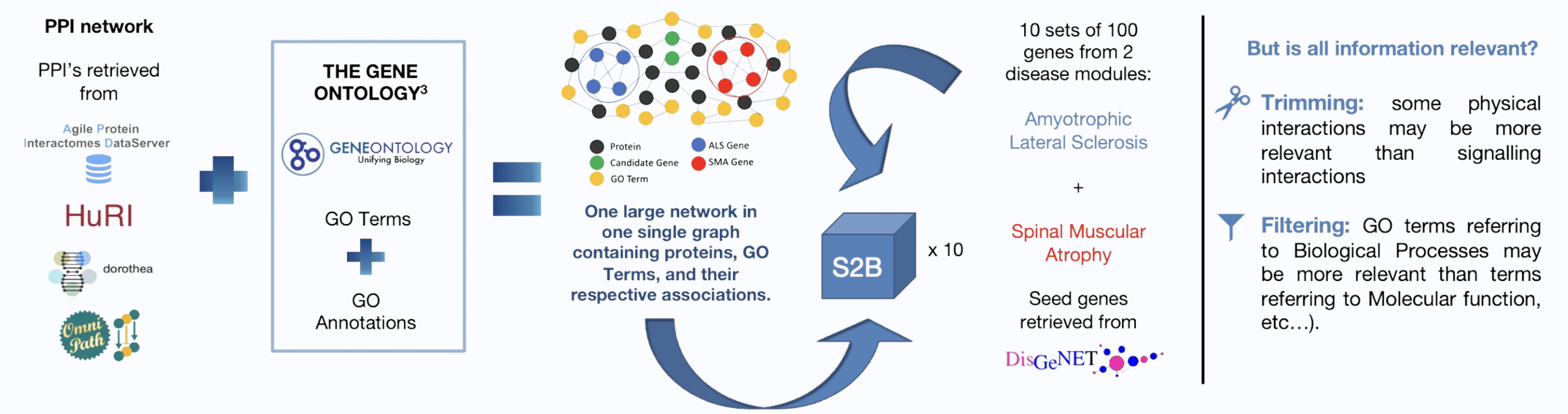
Building a new Network
Assessing the impact of the GO insertions
The introduction of noise
Results
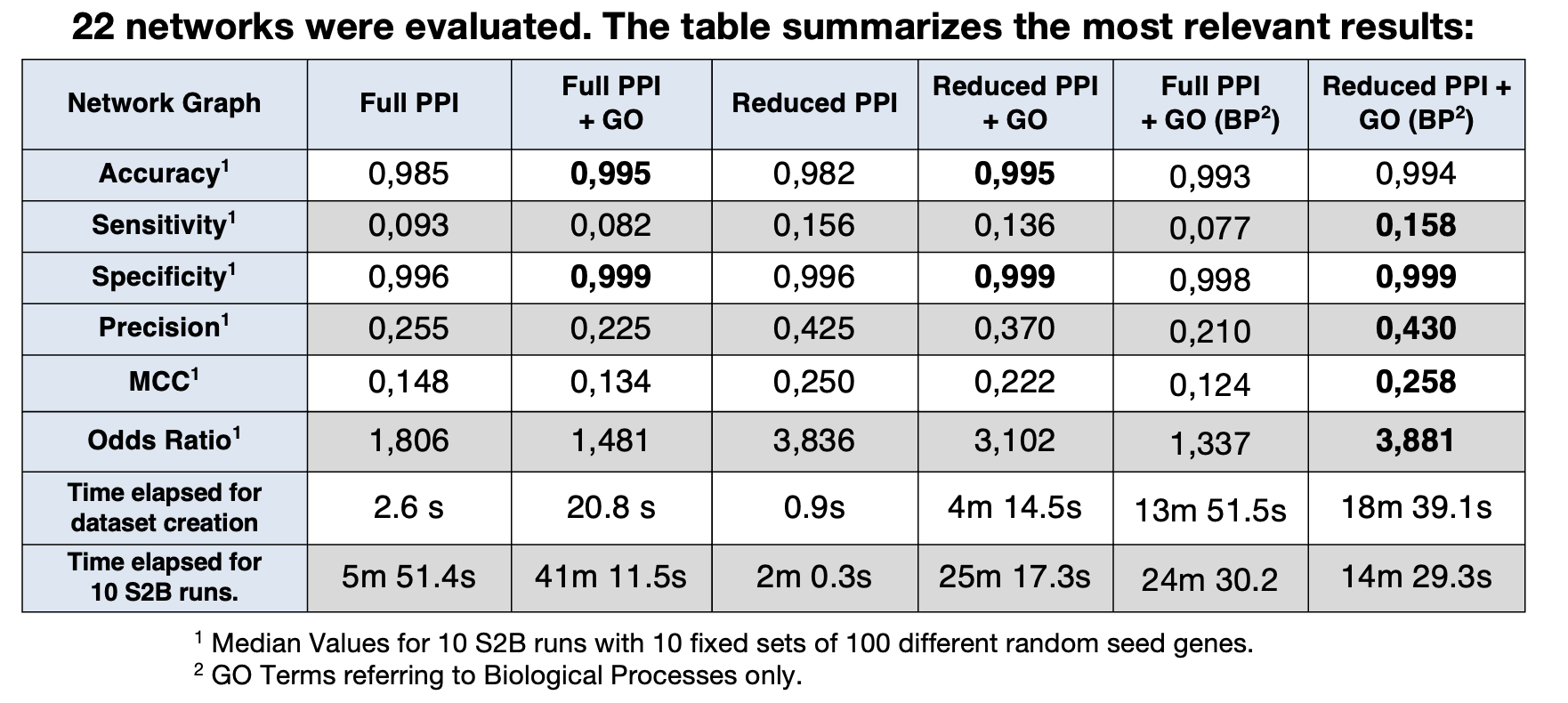
- Our results show that just by itself, the GO doesn't produce a very significant increase in performance to the S2B Method when added to the network.
- However, when using a network with just physical PPI's, we can see a noticeable increase in performance, which shows well the need to trim and filter our data.
- Globally, taking into account performance and run times, the best combination seems to be the one that uses a network containing only physical PPI's, as well as only GO Terms referring to Biological Processes.
What about explainability?
The genes with the highest S2B score are more likely to be in the overlap between disease modules and, hence, more likely to be associated with both diseases.

- Transcription regulation, apoptotic processes and gene expression highly involved in the pathological process of ALS and SMA.
- VCAM1 and TP53 genes encode for proteins related to signal transduction and membrane adhesion.
- These proteins have been highlighted as potential therapeutic candidates to ALS and SMA in experimental studies available in the literature.
Conclusions
- The inclusion of the GO into the PPI network improves S2B’s performance. Filtering and trimming brings further improvement.
- The combination between using only physical PPI’s, and the GO terms referring to biological processes produces the best results yet.
- We also bring more explainability to the method, and are now able to interpret why certain genes are such strong candidates.
- Future work will consist in producing a weighted network. We’ll attribute weights to edges between nodes based on the semantic similarity between their associated GO terms to uncover more relevant interactions.
Funding
This work was supported by the Fundação para a Ciência e a Tecnologia (FCT) under LASIGE Research Unit ref. UIDB/00408/2020 and UIDP/00408/2020, and Partially Supported by FCT Centre grants to BioISI ref. UIDB/04046/2020 and UIDP/04046/2020.






