Motivation
As machine learning is increasingly being used, concerns about data leakage have been raised.
Leakage occurs when information about the target of a data mining problem that should not be legitimately available to mine from is introduced, and it can lead to overestimation of the model’s performance. In biomedical applications, such as protein-protein interaction (PPI) prediction, data leakage can also be an issue since multiple databases and resources reuse the same sources of information.
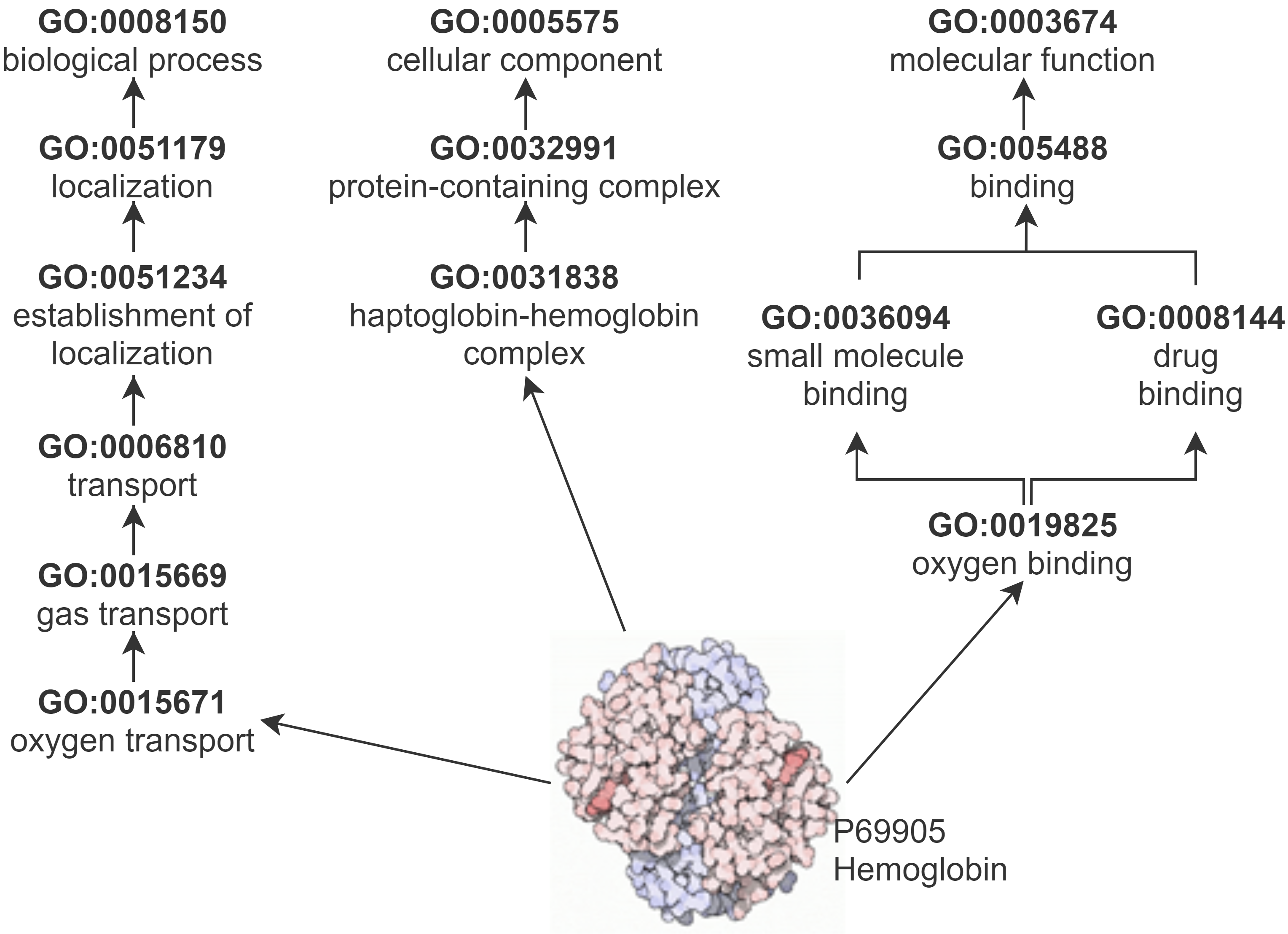
The majority of PPI prediction methods that are based on knowledge graphs (KGs) explore the Gene Ontology (GO) KG, composed of the GO and GO annotations. Since GO annotations can be based on the automated processing of other data sources, the same information that is used to support a PPI in a database to also be used to establish a GO annotation for the proteins.
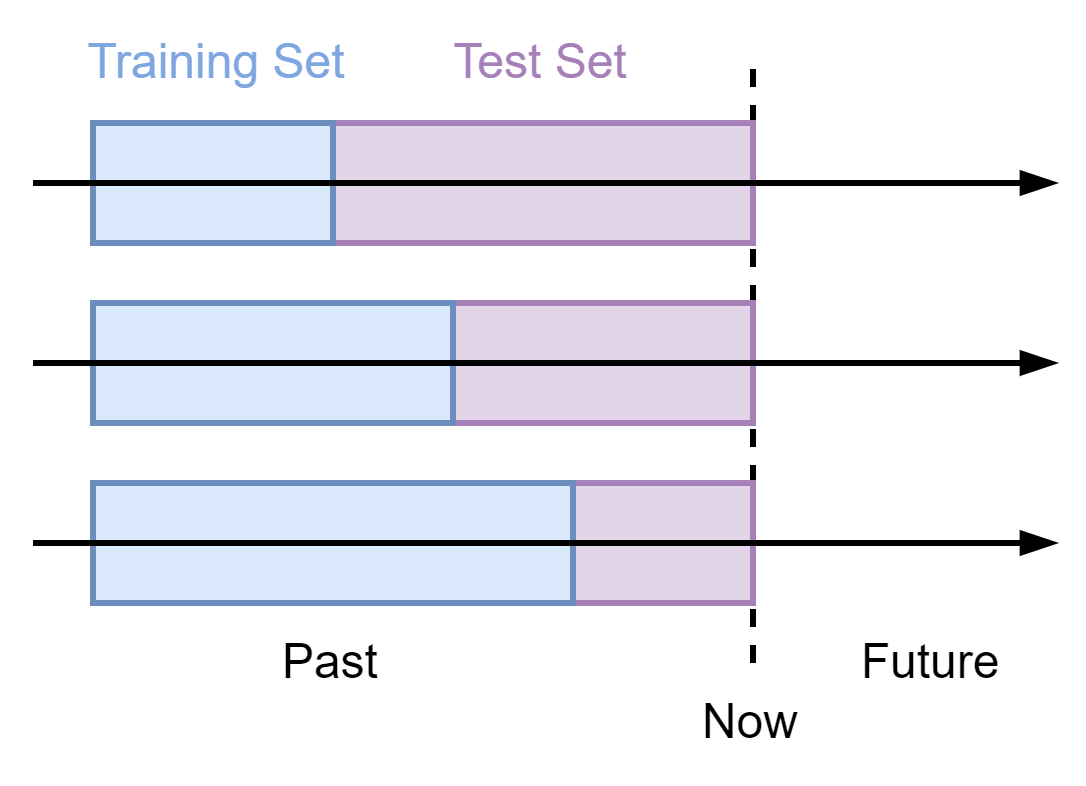
We investigated potential data leakage between the GO KG and the STRING database in the task of PPI prediction, by comparing performance on unseen interactions.
Knowledge Graphs and Data Resources
Gene Ontology Knowledge Graph
GO describes protein function with respect to three semantic aspects: biological processes, cellular components, and molecular functions. GO and the annotations that link proteins to GO classes make up a KG. We obtained archived versions of the GO and GO annotations in 2015, 2017 and 2019 from the GO Data Archive.
Protein-Protein Interaction Datasets
The PPI datasets were obtained from the STRING Database, one of the largest available PPI databases. We built several PPI datasets using three archived versions of the STRING database (v9.1, v10, and v10.5) and the current version (v11). For the current version, we created three datasets each excluding protein pairs present in each of the older versions.
See the number of interactions in each version of the STRING database
We considered the following criteria to select protein pairs from STRING:
(i) each protein must be annotated with the GO;
(ii) protein interactions must be experimentally determined or from curated databases;
(iii) interactions must have a confidence score above 950 to retain only high confidence interactions.
We employed random sampling to create negative pairs composed of the human proteins present in the positive pairs but without any STRING interactions between them, building a balanced dataset.
Protein-Protein Interaction Prediction
Similarity Computation
Employ three KG-based semantic similarity measures to compute semantic similarity: (i) two taxonomic measures (ResnikMax, SimGIC) combined with ICSeco approach; (ii) one based on graph embedding methods (RDF2Vec).
Supervised Learning
Apply six well-known classes of machine learning models to train classifiers: K-Nearest Neighbor (KNN); Genetic Programming (GP); Decision Tree (DT); XGBoost (XGB); Random Forest (RF); Multi-Layer Perceptron (MLP).
Performance Evaluation
For evaluating the quality of a predicted classification, the Weighted Average F-measure (WAF) was used for stratified 10-fold cross-validation. The values reported are the WAFs median.
Results
Conclusions
- The results do not support a clear indication for data bias, indicating that if this problem exists, its magnitude is not affecting the performance of KG-based PPI predictions.
- The results indicate that the relation between the functions of a protein and its interactions do not fundamentally change over time.
Funding
Catia Pesquita, Sara Silva, Rita T. Sousa are funded by the FCT through LASIGE Research Unit, ref. UIDB/00408/2020 and ref. UIDP/00408/2020. Catia Pesquita and Rita T. Sousa are funded by project SMILAX (ref. PTDC/EEI-ESS/4633/2014), Sara Silva by projects BINDER (ref. PTDC/CCI-INF/29168/2017) and PREDICT (ref. PTDC/CCI-CIF/29877/2017), and Rita T. Sousa by FCT PhD grant (ref. SFRH/BD/145377/2019). It was also partially supported by the KATY project which has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 101017453.